Motivation and Background
- Motivation
- What can Protein Structure Prediction do for ML benchmarking?
- How were these datasets generated?
Motivation
In the Machine Learning (ML) field we need benchmarks to evaluate the behaviour and performance of our systems. The test suite is chosen/designed with many characteristics in mind:
- Datasets with diverse numbers of attributes
- Low/High number of classes
- Low/High class inbalance
- High number of instances (scalability purposes)
Real world datasets usually contain noise and inconsistencies, thus being useful to evaluate the robustness of a learning system. However, if our objective is to evaluate the scalability of our system in terms of number of attributes, number of classes, etc. that system is able to cope with, then we need a really broad range of datasets. To achieve this is possible to use some synthetic datasets which we can arbitrarily adjust in dimensions, or we can inflate the real datasets with irrelevant data, introducing a bias to the evaluation procedure.
The datasets in this repository are an alternative family of problems based on real data, that vary the characteristics of the problem in a regular step.
Therefore, the evaluation process could (potentially) be more fair, reliable and robust, since we avoid having to artificially inflate real datasets but at the same time we are using problem far more complicated than toy problems.
What can Protein Structure Prediction do for ML benchmarking?
In short, Protein Structure Prediction (PSP) aims to predict the 3D structure of a protein based on its primary sequence, that is, a chain of amino-acids (a string using a 20-letter alphabet).
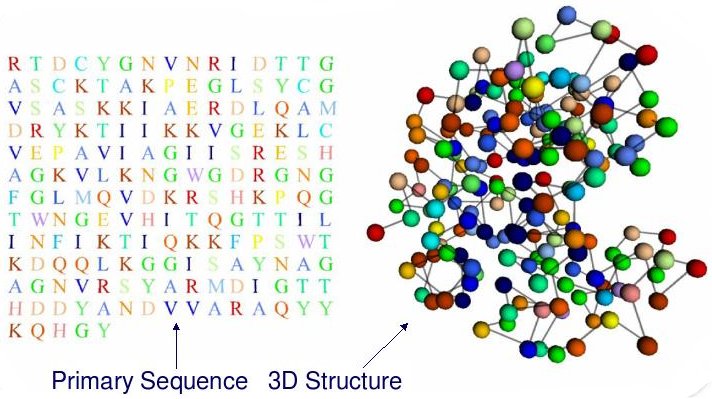
PSP is, overall, an optimization problem. However, each amino-acid can be characterized by several structural features. A good prediction of these features contributes greatly to obtain better models for the 3D PSP problem. These features can be predicted as classification/regression problems.
These features are predicted from the local context (a window of amino acids) of the target in the chain. In the example below, a feature called Coordination Number (CN) for residue i is predicted using information of itself and its two nearest neighbours in the chain sequence, i-1 and i+1.
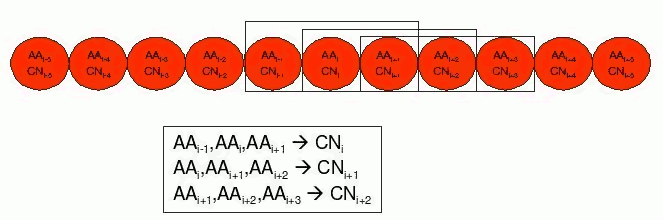
By generating different versions of this problem with different window sizes we can construct a family of datasets of arbitrarily increasing number of attributes, that can be useful to evaluate how a learning system can cope with datasets of different sizes.
Moreover, most of these structural features are defined as continuous variables. Thus suitable to treat them as regression problems. However, it is also usual to discretize them and treat them as classification problems. We can decide the number of bins in which we discretize the feature. Therefore, we can construct too a family of datasets with arbitrarily increasing number of classes. The criteria used to discretize will create datasets with well balanced class distribution (using an uniform-frequency (UF) discretization), or will create datasets with uneven class distribution (using an uniform-length (UL) discretization).
Finally, there are usually two types of basic input information that can be used in these datasets:
- Directly using the amino-acids (AA) of the primary sequence, where one amino acid is defined as one nominal variable with 20 possible values.
- Using a Position-Specific Scoring Matrices (PSSM) representation derived from the primary sequence. The PSSM representation is an statistical profile of the primary sequence that takes into account how this sequence may have evolved. Each amino acid is defined as 20 continuous variables.
As a summary, PSP can provide us with a large variety of ML datasets, derived from trying to predict the same protein structural feature with different formulations of inputs and outputs. Thus, we have an adjustable real-world family of benchmarks suitable for testing the scalability of prediction methods in several fronts.
How were these datasets generated?
- To create the classification datasets we partitioned the CN domain into two, three and five states using both the uniform-width and uniform-frequency discretisation, creating datasets without or with even class distribution, respectively.
- The dataset is derived from a set of 1050 protein chains and ~260000 amino acids (instances) selected using the PDB-REPRDB server.
- Following Kinjo et al., the list of proteins was scrambled 10 times. For each scrambled list the first 950 proteins were used for training. The other 100 proteins for test.
- For each protein, its PDB file, specifying its 3D structure, was downloaded from the Protein Data Bank repository.
- The primary sequence of the protein and the Coordination Number definition of each amino acid were extracted/computed from the PDB file
- The PSSM profiles were computed for each protein from its primary sequence using the PSI-BLAST program using the NR database.
- Given a window size and representation (AA/PSSM) the input data of the training/test sets is constucted using in-house perl scripts. We generated versions of the dataset with a window size ranging from 0 to ±9 amino acids.
- For the regression version of the datasets, the output is directly the CN measure computed previously.
- For the classification datasets, a number of bins and a discretization criteria (balanced/non-balanced classes) is needed. The bins are computed separately for each training set using all of its insances and afterwards applied also to the corresponding test set. We generated datasets with 2 3 and 5 classes.
- Total: 140 versions of the datasets, taking (uncompressed) around 100GB of disk space.
More specific details about the generation of these datasets can be found in:
-
DOI
BibTeX
Coordination number prediction using learning classifier systemsin Proceedings of the 8th annual conference on Genetic and evolutionary computation - GECCO '06, p.247, Seattle, Washington, USA, 2006
@INPROCEEDINGS{Bacardit2006, title = {Coordination number prediction using learning classifier systems}, author = {Bacardit, Jaume and Stout, Michael and Krasnogor, Natalio and Hirst, Jonathan D. and Blazewicz, Jacek}, year = 2006, doi = {10.1145/1143997.1144041}, booktitle = {Proceedings of the 8th annual conference on Genetic and evolutionary computation - GECCO '06}, pages = {247}, address = {Seattle, Washington, USA} } -
DOI
BibTeX
Fast rule representation for continuous attributes in genetics-based machine learningin Proceedings of the 10th annual conference on Genetic and evolutionary computation - GECCO '08, p.1421, Atlanta, GA, USA, 2008
@INPROCEEDINGS{Bacardit2008, title = {Fast rule representation for continuous attributes in genetics-based machine learning}, author = {Bacardit, Jaume and Krasnogor, Natalio}, year = 2008, doi = {10.1145/1389095.1389369}, booktitle = {Proceedings of the 10th annual conference on Genetic and evolutionary computation - GECCO '08}, pages = {1421}, address = {Atlanta, GA, USA} }